
The BackEnd
 |
ONDEX suite
The BackEnd |
|
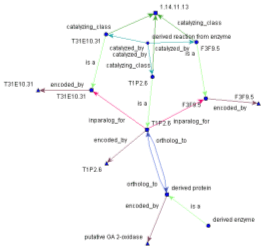
Data Integration Data for integration into the ONDEX system is input from parsed databases including ontologies, pathways, transcription factor, enzyme sources and many others. Various parsers are available to extract data from native formats into the ONDEX graph-based representation. Users can create their own parsers using the ONDEX Parser plugin interface. Data for integration is modelled into a suitable framework of concepts (such as gene, pathway, protein) and relations (such as �belongs_to�, �is_a�) which describe the mapping between concepts. This is performed for each data source. Parsed data is loaded into the Backend and is thoroughly indexed. The ONDEX Backend has facilities to mine out new mappings from the integrated data by looking for evidence of equality, similarity or other relations between concepts from each data source and between each data source. Data concepts are connected to descriptions of the concept (such as name, synonyms) and relations are assigned to relation types or classes. In addition, a more generalised set of data models hold further information about each concept or relation and may contain details of protein, RNA or DNA sequences. The Backend, being based on the Berkeley Java DB is highly scaleable and is able to hold very large quantities of data. It has been developed in pure Java to be platform independent and can serve both local or remote ONDEX Frontend applications. Text Mining The text mining facilities of ONDEX are embedded in the ONDEX Backend. Support is provided for concept based information extraction from free text. Abstract import is supported for the online PubMed system as well as for a local NLM database in XML format. Pre-filtering methods such as positive / negative lists and a date range function are implemented in order to reduce the amount of abstracts to be imported. This will minimise subsequent time extensive operations on the free text. Imported texts will be pre-processed by the NLP techniques word stemming and finally indexed using Lucene. Concept groups may be defined by word lists (species, treatments, etc.) or regular expressions (e.g. for gene symbols). Entities of word lists will be mapped to the integrated concepts in ONDEX in order to derive suitable synonyms and thus expand the concept groups. Co-occurrences of the defined concept groups in the imported texts will be mined and relations will be predicted. Please send comments about this web site to sabr.technical at lists.ondex.org |
  |